Modelling Exercises#
Exercise 1: Model Selection#
Today we are working with the California Housing dataset, which you are already familiar with, as we previously used it while exploring resampling method.
This dataset is based on the 1990 U.S. Census and includes features describing California districts.
Familiarize yourself with the data
What kind of features are in the dataset? What is the target?
Baseline model
Create a baseline linear regression model using all features and evaluate the model through 5-fold cross validation, using R² as the performance metric
Print the individual and average R²
Apply a forward stepwise selection to find a simpler suitable model.
Split the data into 80% training data and 20% testing data (print the shape to confirm it was sucessful)
Perform a forward stepwise selection with a linear regression model, 5-fold CV, R² score, and
parsimoniousfeature selection (refer to documentation for further information)Print the best CV R² as well as the chosen features
Evaluate the model on the test set
import numpy as np
from sklearn.datasets import fetch_california_housing
from mlxtend.feature_selection import SequentialFeatureSelector
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import train_test_split
# 1) Load the California housing dataset
data = fetch_california_housing(as_frame=True)
X = data.data
y = data.target
# 2) Create baseline model
model = LinearRegression()
scores = cross_val_score(model, X, y, cv=5, scoring='r2')
# Print the results
print("R² scores from each fold:", scores)
print("Average R² score:", np.mean(scores))
# 3) Apply a forward stepwise selection
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
print(X_train.shape, X_test.shape)
print(y_train.shape, y_test.shape)
# Forward Sequential Feature Selector
sfs_forward = SequentialFeatureSelector(
estimator=LinearRegression(),
k_features="parsimonious",
forward=True,
floating=False,
scoring='r2',
cv=5,
verbose=0)
sfs_forward.fit(X_train, y_train)
print(f">> Forward SFS:")
print(f" Best CV R² : {sfs_forward.k_score_:.3f}")
print(f" Optimal # feats : {len(sfs_forward.k_feature_idx_)}")
print(f" Feature names : {sfs_forward.k_feature_names_}")
# 4) Evaluate the model
selected_features = list(sfs_forward.k_feature_names_)
X_train_selected = X_train[selected_features]
X_test_selected = X_test[selected_features]
# Train and evaluate
model.fit(X_train_selected, y_train)
test_r2 = model.score(X_test_selected, y_test)
print(f"Test R² for the sfs model: {test_r2:.4f}")
R² scores from each fold: [0.54866323 0.46820691 0.55078434 0.53698703 0.66051406]
Average R² score: 0.5530311140279571
(16512, 8) (4128, 8)
(16512,) (4128,)
>> Forward SFS:
Best CV R² : 0.612
Optimal # feats : 7
Feature names : ('MedInc', 'HouseAge', 'AveRooms', 'AveBedrms', 'AveOccup', 'Latitude', 'Longitude')
Test R² for the sfs model: 0.5757
Exercise 2: LASSO#
Please implement a Lasso regression model similar to the Ridge model in the Regularization section.
import pandas as pd
import numpy as np
import statsmodels.api as sm
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LassoCV
from sklearn.model_selection import train_test_split
# Data related processing
hitters = sm.datasets.get_rdataset("Hitters", "ISLR").data
hitters_subset = hitters[["Salary", "AtBat", "Runs","RBI", "CHits", "CAtBat", "CRuns", "CWalks", "Assists", "Hits", "HmRun", "Years", "Errors", "Walks"]].copy()
hitters_subset = hitters_subset.drop(columns=["CRuns", "CAtBat"]) # Remove highly correlated features (see previous session)
hitters_subset.dropna(inplace=True) # drop rows containing missing data
y = hitters_subset["Salary"]
X = hitters_subset.drop(columns=["Salary"])
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=42)
scaler = StandardScaler() # Scale predictors to mean=0 and std=1
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
# Lasso
lambda_range = np.linspace(0.001, 20, 100)
# Get the optimal lambda
lasso_cv = LassoCV(alphas=lambda_range)
lasso_cv.fit(X_train_scaled, y_train)
print(f"Optimal alpha: {lasso_cv.alpha_}\n")
# Get training R²
train_score_ridge= lasso_cv.score(X_train_scaled, y_train)
print(f"Training R²: {train_score_ridge}\n")
# Put the coefficients into a nicely formatted df for visualization
coef_table = pd.DataFrame({
'Predictor': X_train.columns,
'Beta': lasso_cv.coef_
})
coef_table = coef_table.reindex(coef_table['Beta'].abs().sort_values(ascending=False).index)
print(coef_table, "\n")
test_score_ridge= lasso_cv.score(X_test_scaled, y_test)
print(f"Test R²: {test_score_ridge}")
Optimal alpha: 20.0
Training R²: 0.47908195299121104
Predictor Beta
3 CHits 177.984173
6 Hits 101.982447
7 HmRun 52.177420
10 Walks 41.664953
2 RBI 0.000000
0 AtBat 0.000000
1 Runs 0.000000
5 Assists -0.000000
4 CWalks 0.000000
8 Years 0.000000
9 Errors -0.000000
Test R²: 0.31479649243077035
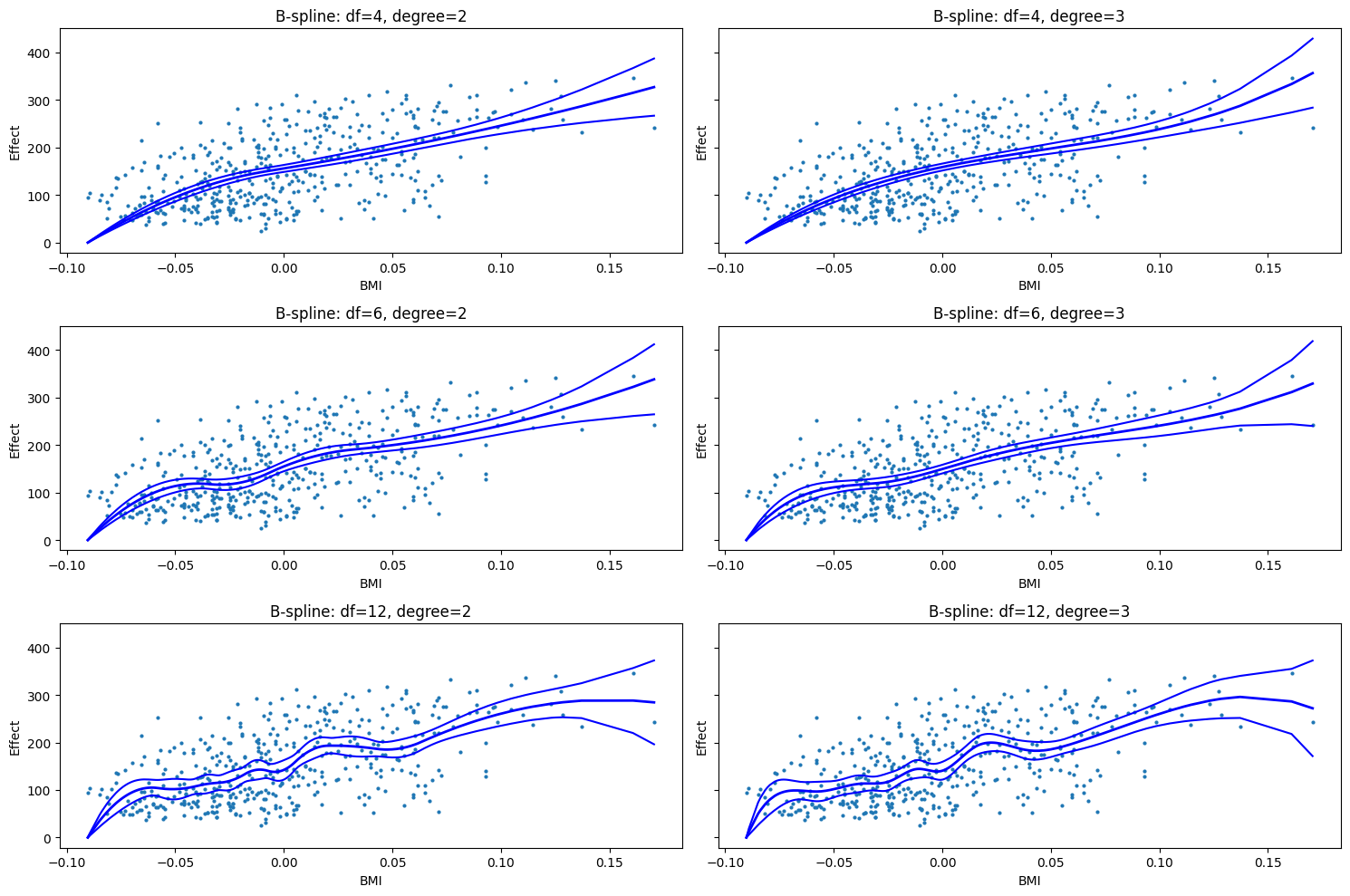
Exercise 3: GAMs (1)#
Objective: Understand how the number of basis functions (df) and the polynomial degree (degree) affect the flexibility of a spline and the resulting fit in a Generalized Additive Model.
Use the diabetes dataset and focus on the relationship between
bmiandtarget.We want to test different combinations of parameters. For the dfs, please use 4, 6, 12. For the degree, please use 2 and 3 (quadratic and cubic).
Fit the GAMs for each parameter combination. The resulting models will be plotted automatically for visual comparison.
import matplotlib.pyplot as plt
from sklearn.datasets import load_diabetes
from statsmodels.gam.api import GLMGam, BSplines
# 1. Get bmi as x and the target as y
data = load_diabetes(as_frame=True)
x = data.data[['bmi']]
y = data.target
# 2. Define possible parameters
df_values = [4, 6, 12]
degree_values = [2, 3]
# 3. PLot partial effect for each combination of df and degree
fig, axes = plt.subplots(len(df_values), len(degree_values), figsize=(15, 10), sharey=True)
for i, df_val in enumerate(df_values):
for j, deg_val in enumerate(degree_values):
bs = BSplines(x, df=df_val, degree=deg_val)
gam = GLMGam(y, smoother=bs)
res = gam.fit()
res.plot_partial(0, cpr=True, ax=axes[i, j])
axes[i, j].set_title(f'B-spline: df={df_val}, degree={deg_val}')
axes[i, j].set_xlabel('BMI')
axes[i, j].set_ylabel('Effect')
plt.tight_layout()
plt.show()
Exercise 4: GAMs (2)#
We now use the wage dataset, which contains income information for a group of workers, along with demographic and employment-related features such as age, education, marital status, and job class.
Explore the dataset
Which variables are numeric?
Which ones are categorical?
Fit a GAM predicting
wagefromage,year,education,jobclass, andmaritl
Note: For categorical features we use a one-hot encoding with pd.get_dummies()
import pandas as pd
from ISLP import load_data
from statsmodels.gam.api import GLMGam, BSplines
# Load data
Wage = load_data('Wage')
# Continuous features
spline_features = ['age', 'year']
X_spline = Wage[spline_features]
# Categorical features — one-hot encode
categoricals = ['education', 'jobclass', 'maritl']
X_cat = pd.get_dummies(Wage[categoricals], drop_first=True)
# Outcome
y = Wage['wage']
# Create BSpline basis
bs = BSplines(X_spline, df=[6]*len(spline_features), degree=[3]*len(spline_features))
# Fit GAM
gam = GLMGam(y, exog=X_cat, smoother=bs)
res = gam.fit()
print(res.summary())
Generalized Linear Model Regression Results
==============================================================================
Dep. Variable: wage No. Observations: 3000
Model: GLMGam Df Residuals: 2981
Model Family: Gaussian Df Model: 18.00
Link Function: Identity Scale: 1230.5
Method: PIRLS Log-Likelihood: -14920.
Date: Mon, 23 Jun 2025 Deviance: 3.6680e+06
Time: 12:44:09 Pearson chi2: 3.67e+06
No. Iterations: 3 Pseudo R-squ. (CS): 0.3436
Covariance Type: nonrobust
================================================================================================
coef std err z P>|z| [0.025 0.975]
------------------------------------------------------------------------------------------------
education_2. HS Grad 16.2973 2.337 6.975 0.000 11.718 20.877
education_3. Some College 27.8392 2.505 11.114 0.000 22.930 32.749
education_4. College Grad 41.0763 2.533 16.219 0.000 36.113 46.040
education_5. Advanced Degree 63.9348 2.792 22.897 0.000 58.462 69.408
jobclass_2. Information 4.8618 1.355 3.589 0.000 2.206 7.517
maritl_2. Married 13.3632 1.875 7.128 0.000 9.689 17.038
maritl_3. Widowed -0.2441 8.279 -0.029 0.976 -16.470 15.982
maritl_4. Divorced -0.1040 3.039 -0.034 0.973 -6.059 5.851
maritl_5. Separated 7.0577 5.046 1.399 0.162 -2.832 16.947
age_s0 63.8878 5.262 12.142 0.000 53.575 74.200
age_s1 63.1018 4.154 15.191 0.000 54.960 71.243
age_s2 74.8020 5.531 13.524 0.000 63.961 85.643
age_s3 56.5913 8.534 6.631 0.000 39.865 73.318
age_s4 47.4594 12.372 3.836 0.000 23.211 71.708
year_s0 4.3025 4.127 1.043 0.297 -3.786 12.391
year_s1 7.1829 4.559 1.575 0.115 -1.753 16.119
year_s2 10.0632 4.781 2.105 0.035 0.692 19.435
year_s3 7.1947 4.284 1.679 0.093 -1.202 15.591
year_s4 10.7707 2.344 4.595 0.000 6.176 15.365
================================================================================================
Exercise 5: KNN#
You will implement a K-Nearest Neighbours (KNN) classifier to predict whether a patient is likely to have malignant or benign breast cancer based on several features. The data is already loaded for you, but please have a look a the documentation to quickly refresh you memory about the dataset.
import pandas as pd
from sklearn.datasets import load_breast_cancer
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, classification_report
from sklearn.preprocessing import StandardScaler
from sklearn.neighbors import KNeighborsClassifier
# Load the data
data = load_breast_cancer()
X, y = data.data, data.target
# Create a DataFrame for easier inspection and manipulation
df = pd.DataFrame(X, columns=data.feature_names)
df['target'] = y
df
| mean radius | mean texture | mean perimeter | mean area | mean smoothness | mean compactness | mean concavity | mean concave points | mean symmetry | mean fractal dimension | ... | worst texture | worst perimeter | worst area | worst smoothness | worst compactness | worst concavity | worst concave points | worst symmetry | worst fractal dimension | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 17.99 | 10.38 | 122.80 | 1001.0 | 0.11840 | 0.27760 | 0.30010 | 0.14710 | 0.2419 | 0.07871 | ... | 17.33 | 184.60 | 2019.0 | 0.16220 | 0.66560 | 0.7119 | 0.2654 | 0.4601 | 0.11890 | 0 |
| 1 | 20.57 | 17.77 | 132.90 | 1326.0 | 0.08474 | 0.07864 | 0.08690 | 0.07017 | 0.1812 | 0.05667 | ... | 23.41 | 158.80 | 1956.0 | 0.12380 | 0.18660 | 0.2416 | 0.1860 | 0.2750 | 0.08902 | 0 |
| 2 | 19.69 | 21.25 | 130.00 | 1203.0 | 0.10960 | 0.15990 | 0.19740 | 0.12790 | 0.2069 | 0.05999 | ... | 25.53 | 152.50 | 1709.0 | 0.14440 | 0.42450 | 0.4504 | 0.2430 | 0.3613 | 0.08758 | 0 |
| 3 | 11.42 | 20.38 | 77.58 | 386.1 | 0.14250 | 0.28390 | 0.24140 | 0.10520 | 0.2597 | 0.09744 | ... | 26.50 | 98.87 | 567.7 | 0.20980 | 0.86630 | 0.6869 | 0.2575 | 0.6638 | 0.17300 | 0 |
| 4 | 20.29 | 14.34 | 135.10 | 1297.0 | 0.10030 | 0.13280 | 0.19800 | 0.10430 | 0.1809 | 0.05883 | ... | 16.67 | 152.20 | 1575.0 | 0.13740 | 0.20500 | 0.4000 | 0.1625 | 0.2364 | 0.07678 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 564 | 21.56 | 22.39 | 142.00 | 1479.0 | 0.11100 | 0.11590 | 0.24390 | 0.13890 | 0.1726 | 0.05623 | ... | 26.40 | 166.10 | 2027.0 | 0.14100 | 0.21130 | 0.4107 | 0.2216 | 0.2060 | 0.07115 | 0 |
| 565 | 20.13 | 28.25 | 131.20 | 1261.0 | 0.09780 | 0.10340 | 0.14400 | 0.09791 | 0.1752 | 0.05533 | ... | 38.25 | 155.00 | 1731.0 | 0.11660 | 0.19220 | 0.3215 | 0.1628 | 0.2572 | 0.06637 | 0 |
| 566 | 16.60 | 28.08 | 108.30 | 858.1 | 0.08455 | 0.10230 | 0.09251 | 0.05302 | 0.1590 | 0.05648 | ... | 34.12 | 126.70 | 1124.0 | 0.11390 | 0.30940 | 0.3403 | 0.1418 | 0.2218 | 0.07820 | 0 |
| 567 | 20.60 | 29.33 | 140.10 | 1265.0 | 0.11780 | 0.27700 | 0.35140 | 0.15200 | 0.2397 | 0.07016 | ... | 39.42 | 184.60 | 1821.0 | 0.16500 | 0.86810 | 0.9387 | 0.2650 | 0.4087 | 0.12400 | 0 |
| 568 | 7.76 | 24.54 | 47.92 | 181.0 | 0.05263 | 0.04362 | 0.00000 | 0.00000 | 0.1587 | 0.05884 | ... | 30.37 | 59.16 | 268.6 | 0.08996 | 0.06444 | 0.0000 | 0.0000 | 0.2871 | 0.07039 | 1 |
569 rows × 31 columns
Please implement the following:
Subset the dataframe to use
mean area,mean radius, andmean smoothnessas features (X), andtargetas the target (y)Scale the predictors to mean 0 and variance 1
Split the data into a training and a testing set (70/30)
Train a kNN classifier with \(k=5\)
Evaluate the performance for the testing set. Please use the
accuracy_score()as well as theclassification_report()function.
# Select features and target
X = df[['mean area', 'mean radius', 'mean smoothness']]
y = df['target']
# Scale the features
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
# Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(X_scaled, y, test_size=0.2, random_state=42)
# Perform KNN classification
knn = KNeighborsClassifier(n_neighbors=5)
knn.fit(X_train, y_train)
# Get predictions
y_pred = knn.predict(X_test)
print("Accuracy:", accuracy_score(y_test, y_pred))
print("\nClassification Report:\n", classification_report(y_test, y_pred))
Accuracy: 0.8947368421052632
Classification Report:
precision recall f1-score support
0 0.88 0.84 0.86 43
1 0.90 0.93 0.92 71
accuracy 0.89 114
macro avg 0.89 0.88 0.89 114
weighted avg 0.89 0.89 0.89 114
The classification model from the previous step has two main limitations:
It is trained and evaluated on a single data split
It uses a single \(k\) even though we do not know if it is optimal
Please do the following:
Implement 5-fold cross validation
Train models for \(k\) ranging from 1 to 200 and plot the mean accuracy over all folds
from sklearn.model_selection import cross_val_score
import seaborn as sns
k_values = range(1, 200)
mean_accuracies = []
# 5-fold cross-validation for different k values
for k in k_values:
knn = KNeighborsClassifier(n_neighbors=k)
scores = cross_val_score(knn, X_scaled, y, cv=5)
mean_accuracies.append(scores.mean())
# Plot
fig, ax = plt.subplots()
sns.set_theme(style="whitegrid")
sns.lineplot(x=k_values, y=mean_accuracies, linestyle='-', ax=ax)
ax.set(xlabel='Number of neighbors (k)', ylabel='Mean accuracy', title='Mean accuracy for different k');
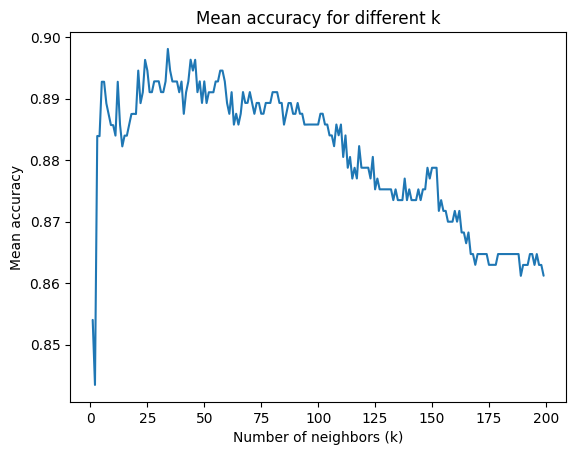
Discuss the results. How is the performance in general? Which \(k\) would you chose?
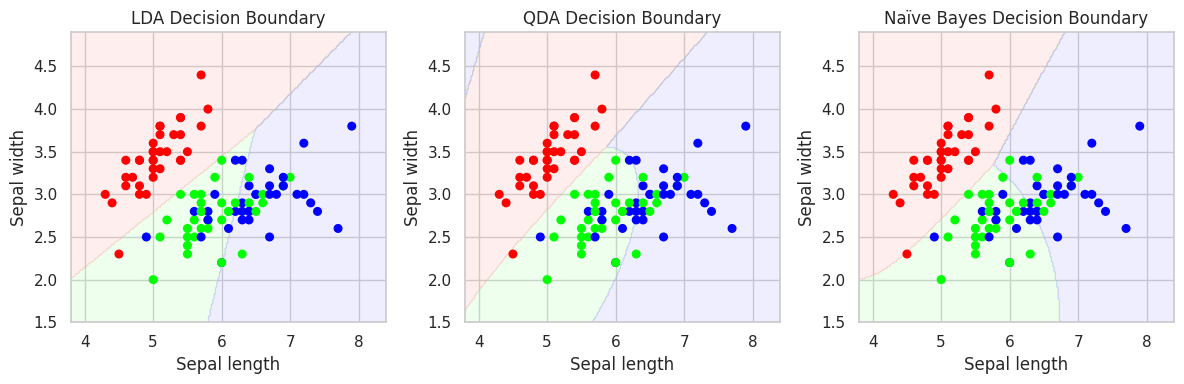
Exercise 6: LDA, QDA & Naïve Bayes#
Once again, we will use the Iris dataset for classificationa analysis. Your task is to compare the performance of LDA, QDA, and Gaussian Naïve Bayes!
Load the
irisdataset fromsklearn.datasets. We will use only the first two features (sepal length and width)TODO:Split the data into training and test sets (use stratification!)TODO:Fit LDA, QDA, and Naïve Bayes classifiers to the training data and orint the classification report for all models on the test dataPlot the decision boundaries for both models
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import load_iris
from sklearn.metrics import classification_report
from sklearn.model_selection import train_test_split
from matplotlib.colors import ListedColormap
# 1. Load data
iris = load_iris()
X = iris.data[:, :2]
y = iris.target
target_names = iris.target_names
# 2. Split into train/test
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, stratify=y, random_state=123)
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
# 3. TODO: Fit a LDA model and print the classification report
lda = LinearDiscriminantAnalysis()
lda.fit(X_train, y_train)
print(classification_report(y_test, lda.predict(X_test)))
precision recall f1-score support
0 1.00 1.00 1.00 15
1 0.69 0.60 0.64 15
2 0.65 0.73 0.69 15
accuracy 0.78 45
macro avg 0.78 0.78 0.78 45
weighted avg 0.78 0.78 0.78 45
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis
# 3. TODO: Fit a QDA model and print the classification report
qda = QuadraticDiscriminantAnalysis()
qda.fit(X_train, y_train)
print(classification_report(y_test, qda.predict(X_test)))
precision recall f1-score support
0 1.00 1.00 1.00 15
1 0.73 0.53 0.62 15
2 0.63 0.80 0.71 15
accuracy 0.78 45
macro avg 0.79 0.78 0.77 45
weighted avg 0.79 0.78 0.77 45
from sklearn.naive_bayes import GaussianNB
# 3. TODO: Fit a Gaussian Naive Bayes model and print the classification report
gnb = GaussianNB()
gnb.fit(X_train, y_train)
print(classification_report(y_test, gnb.predict(X_test)))
precision recall f1-score support
0 1.00 1.00 1.00 15
1 0.71 0.67 0.69 15
2 0.69 0.73 0.71 15
accuracy 0.80 45
macro avg 0.80 0.80 0.80 45
weighted avg 0.80 0.80 0.80 45
# 4. Plot the decision boundaries for all 3 classifiers
# Plotting function
def plot_decision_boundary(model, X, y, title, ax):
h = .02
x_min, x_max = X[:, 0].min() - .5, X[:, 0].max() + .5
y_min, y_max = X[:, 1].min() - .5, X[:, 1].max() + .5
xx, yy = np.meshgrid(np.arange(x_min, x_max, h),
np.arange(y_min, y_max, h))
Z = model.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
cmap_light = ListedColormap(['#FFAAAA', '#AAFFAA', '#AAAAFF'])
cmap_bold = ListedColormap(['#FF0000', '#00FF00', '#0000FF'])
ax.contourf(xx, yy, Z, cmap=cmap_light, alpha=0.2)
scatter = ax.scatter(X[:, 0], X[:, 1], c=y, cmap=cmap_bold, s=30)
ax.set_xlim(xx.min(), xx.max())
ax.set_ylim(yy.min(), yy.max())
ax.set_title(title)
ax.set_xlabel('Sepal length')
ax.set_ylabel('Sepal width')
# Create plots for all 3 classifiers
fig, axes = plt.subplots(1, 3, figsize=(12, 4))
plot_decision_boundary(lda, X_train, y_train, "LDA Decision Boundary", axes[0])
plot_decision_boundary(qda, X_train, y_train, "QDA Decision Boundary", axes[1])
plot_decision_boundary(gnb, X_train, y_train, "Naïve Bayes Decision Boundary", axes[2])
plt.tight_layout()
Exercise 7: Trees#
Inspect the data
How many features are there and what are they?
What is the target?
Split the data into a train and test set, and make sure the classes are equally distributed (
stratify=y)Fit the DecisionTreeClassifier(max_depth=3) and report train vs. test accuracy.
Tree inspection (discuss in group)
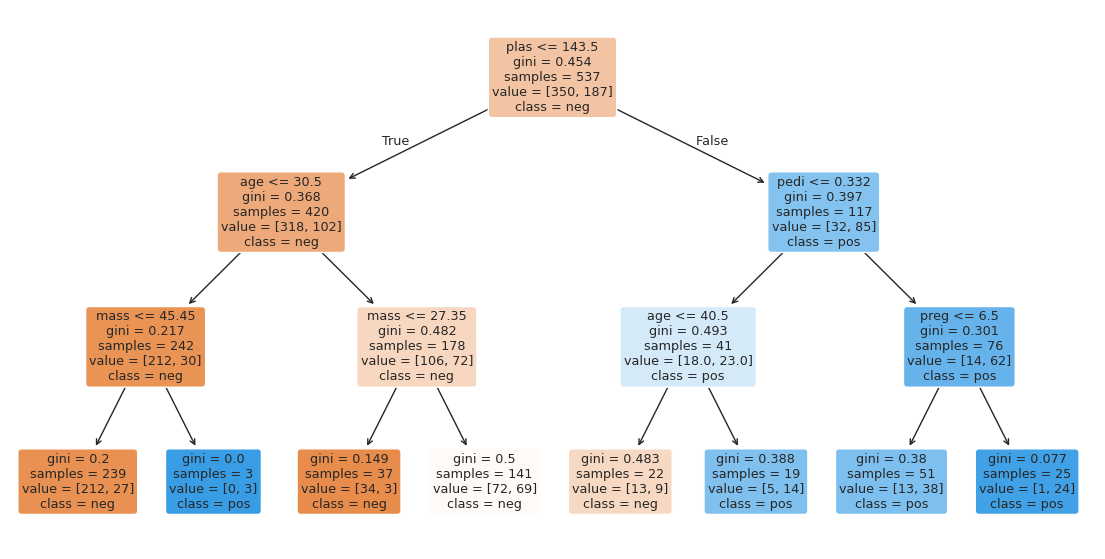
After fitting the model, the tree will be plotted automatically
What is the very first split (feature name and threshold)?
Which leaf nodes are pure, and which have mixed classes?
from sklearn.datasets import fetch_openml
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier, plot_tree
from sklearn.metrics import accuracy_score
import matplotlib.pyplot as plt
# 1) Load and inspect data
diab = fetch_openml("diabetes", version=1, as_frame=True)
X = diab.data
y = diab.target
print(X.head(), "\n")
print(y.head())
# 2) Split data
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, stratify=y, random_state=0)
# 3) Fit tree
clf = DecisionTreeClassifier(max_depth=3, random_state=0)
clf.fit(X_train, y_train)
print("\nTrain accuracy:", accuracy_score(y_train, clf.predict(X_train)))
print("Test accuracy: ", accuracy_score(y_test, clf.predict(X_test)))
# 5) Plot tree
plt.figure(figsize=(14,7))
plot_tree(clf, feature_names=X.columns, class_names=["neg","pos"], filled=True, rounded=True);
preg plas pres skin insu mass pedi age
0 6 148 72 35 0 33.6 0.627 50
1 1 85 66 29 0 26.6 0.351 31
2 8 183 64 0 0 23.3 0.672 32
3 1 89 66 23 94 28.1 0.167 21
4 0 137 40 35 168 43.1 2.288 33
0 tested_positive
1 tested_negative
2 tested_positive
3 tested_negative
4 tested_positive
Name: class, dtype: category
Categories (2, object): ['tested_negative', 'tested_positive']
Train accuracy: 0.7635009310986964
Test accuracy: 0.7445887445887446
Let’s see if we can improve the classification performance with a random forest classifier and hyperparameter tuning!
Set up the clasifier + a parameter grid for grid search with 5-fold CV
n_estimators: 50, 100, 200
max_depth: None, 10, 20
min_samples_split: 2, 5, 10
max_features: “sqrt”, “log2”, 0.5
Fit the model with the grid search
Print the best hyperparameters
Evaluate the best model on the test set
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import accuracy_score, classification_report
# 1) Set up Random Forest + parameter grid
rf = RandomForestClassifier(random_state=0)
param_grid = {
'n_estimators': [50, 100, 200],
'max_depth': [None, 10, 20],
'min_samples_split': [2, 5, 10],
'max_features': ['sqrt', 'log2', 0.5]
}
# 2) Fit on training data
grid = GridSearchCV(rf, param_grid=param_grid, cv=5, scoring='accuracy', verbose=1)
grid.fit(X_train, y_train)
# 3) Print best hyperparameters
print("Best parameters:", grid.best_params_)
print(f"CV accuracy: {grid.best_score_:.3f}")
# 4) Evaluate on the held‐out test set
best_rf = grid.best_estimator_
y_pred = best_rf.predict(X_test)
print(f"\nTest accuracy: {accuracy_score(y_test, y_pred):.3f}\n")
print("Classification Report:")
print(classification_report(y_test, y_pred, target_names=['neg','pos']))
Fitting 5 folds for each of 81 candidates, totalling 405 fits
Best parameters: {'max_depth': 10, 'max_features': 'sqrt', 'min_samples_split': 2, 'n_estimators': 50}
CV accuracy: 0.773
Test accuracy: 0.788
Classification Report:
precision recall f1-score support
neg 0.83 0.85 0.84 150
pos 0.71 0.67 0.69 81
accuracy 0.79 231
macro avg 0.77 0.76 0.76 231
weighted avg 0.79 0.79 0.79 231
/home/mibur/miniconda3/envs/psy111/lib/python3.13/site-packages/numpy/ma/core.py:2881: RuntimeWarning: invalid value encountered in cast
_data = np.array(data, dtype=dtype, copy=copy,
Exercise 8: SVC#
For the SVC exercise we will use the fmri dataset from seaborn, which contains measurements of brain activity (signal) in two brain regions (frontal and parietal) under two event types (stim vs. cue).
import seaborn as sns
df = sns.load_dataset("fmri")
df
| subject | timepoint | event | region | signal | |
|---|---|---|---|---|---|
| 0 | s13 | 18 | stim | parietal | -0.017552 |
| 1 | s5 | 14 | stim | parietal | -0.080883 |
| 2 | s12 | 18 | stim | parietal | -0.081033 |
| 3 | s11 | 18 | stim | parietal | -0.046134 |
| 4 | s10 | 18 | stim | parietal | -0.037970 |
| ... | ... | ... | ... | ... | ... |
| 1059 | s0 | 8 | cue | frontal | 0.018165 |
| 1060 | s13 | 7 | cue | frontal | -0.029130 |
| 1061 | s12 | 7 | cue | frontal | -0.004939 |
| 1062 | s11 | 7 | cue | frontal | -0.025367 |
| 1063 | s0 | 0 | cue | parietal | -0.006899 |
1064 rows × 5 columns
We will try to answer a very simple research question:
Can we distinguish between
cueandstimevents based on the fMRI signal in theparietalandfrontalbrain regions?
To do this, we need to turn the long‐format data into a classic “feature matrix” (one row = one sample, two columns = our two brain‐region signals) plus a corresponding label vector (cue/stim):
df_wide = df.pivot_table(
index=["subject","timepoint","event"],
columns="region",
values="signal"
).reset_index()
df_wide.columns.name = None
X = df_wide[["frontal","parietal"]]
y = df_wide["event"].map({"cue":0,"stim":1})
print("\nFeatures:")
print(X)
print("\nTarget:")
print(y)
Features:
frontal parietal
0 0.007766 -0.006899
1 -0.021452 -0.039327
2 0.016440 0.000300
3 -0.021054 -0.035735
4 0.024296 0.033220
.. ... ...
527 -0.036739 -0.131641
528 -0.004900 -0.036362
529 -0.030099 -0.121574
530 -0.000643 -0.051040
531 -0.009959 -0.103513
[532 rows x 2 columns]
Target:
0 0
1 1
2 0
3 1
4 0
..
527 1
528 0
529 1
530 0
531 1
Name: event, Length: 532, dtype: int64
With the features and target in the correct form, please perform the following tasks:
Split the data into a train and test set
Scale the predictors to mean 0 and std 1
Fit a linear as well as a rbf SVC and discuss the classification reports
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.svm import SVC
from sklearn.metrics import classification_report
# 1. Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split( X, y, test_size=0.3, stratify=y, random_state=42)
# 2. Scale the features after splitting (important to avoid data leakage)
scaler = StandardScaler()
X_train_sc = scaler.fit_transform(X_train)
X_test_sc = scaler.transform(X_test)
# 3. Fit the SVC models and compare the classification reports
clf_lin = SVC(kernel="linear", random_state=42)
clf_lin.fit(X_train_sc, y_train)
y_pred_lin = clf_lin.predict(X_test_sc)
print("Linear SVC\n", classification_report(y_test, y_pred_lin))
clf_rbf = SVC(kernel="rbf", random_state=42)
clf_rbf.fit(X_train_sc, y_train)
y_pred_rbf = clf_rbf.predict(X_test_sc)
print("RBF SVC\n", classification_report(y_test, y_pred_rbf))
Linear SVC
precision recall f1-score support
0 0.55 0.99 0.71 80
1 0.94 0.20 0.33 80
accuracy 0.59 160
macro avg 0.75 0.59 0.52 160
weighted avg 0.75 0.59 0.52 160
RBF SVC
precision recall f1-score support
0 0.67 0.85 0.75 80
1 0.79 0.57 0.67 80
accuracy 0.71 160
macro avg 0.73 0.71 0.71 160
weighted avg 0.73 0.71 0.71 160
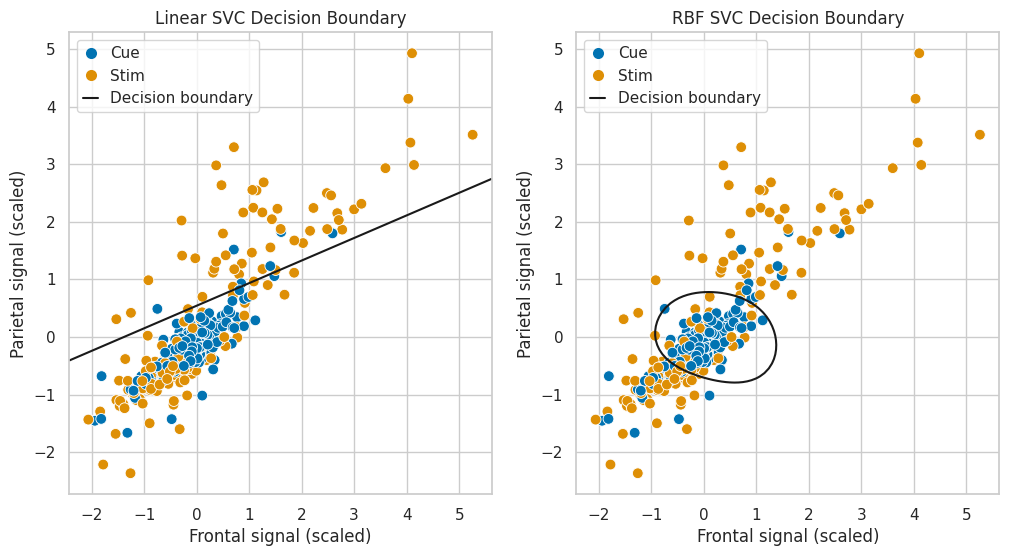
After fitting both models, you can run the code chunk below to plot the decision boundary:
import matplotlib.pyplot as plt
from matplotlib.lines import Line2D
def plot_svc_decision_function(model, ax=None):
"""Plot the decision boundary for a trained 2D SVC model."""
# Set up grid
xlim = ax.get_xlim()
ylim = ax.get_ylim()
xx, yy = np.meshgrid(np.linspace(*xlim, 100), np.linspace(*ylim, 100))
grid = np.c_[xx.ravel(), yy.ravel()]
decision_values = model.decision_function(grid).reshape(xx.shape)
ax.contour(xx, yy, decision_values, levels=[0], linestyles=['-'], colors='k')
# Plot
fig, ax = plt.subplots(1,2, figsize=(12, 6))
legend_elements = [
Line2D([0], [0], marker='o', linestyle='None', markersize=8, label='Cue', markerfacecolor="#0173B2", markeredgecolor='None'),
Line2D([0], [0], marker='o', linestyle='None', markersize=8, label='Stim', markerfacecolor="#DE8F05", markeredgecolor='None'),
Line2D([0], [0], color='k', linestyle='-', label='Decision boundary')]
# Linear SVC
sns.scatterplot(x = X_train_sc[:, 0], y = X_train_sc[:, 1], hue = y_train.map({0:"cue",1:"stim"}), palette = ["#0173B2", "#DE8F05"], s = 60, ax = ax[0], legend=None)
ax[0].set(xlabel = "Frontal signal (scaled)", ylabel = "Parietal signal (scaled)", title = "Linear SVC Decision Boundary")
plot_svc_decision_function(clf_lin, ax=ax[0])
ax[0].legend(handles=legend_elements, loc="upper left", handlelength=1)
# RBF SVC
sns.scatterplot(x = X_train_sc[:, 0], y = X_train_sc[:, 1], hue = y_train.map({0:"cue",1:"stim"}), palette = ["#0173B2", "#DE8F05"], s = 60, ax = ax[1], legend=None)
ax[1].set(xlabel = "Frontal signal (scaled)", ylabel = "Parietal signal (scaled)", title = "RBF SVC Decision Boundary")
plot_svc_decision_function(clf_rbf, ax=ax[1])
ax[1].legend(handles=legend_elements, loc="upper left", handlelength=1);
Training a SVC on more complex datasets usually requires a parameter search to find the optimal hyperparameters. Please implement a grid search with the following options:
Kernel: rbf
C: np.logspace(-2,2,5)
gamma: np.logspace(-3,1,5)
cv: 5-fold
scoring: accuracy
Print the optimal parameters and the corresponding accuracies for taining and testing.
from sklearn.model_selection import GridSearchCV
param_grid = {
"C": np.logspace(-2,2,5),
"gamma": np.logspace(-3,1,5)
}
grid = GridSearchCV(SVC(kernel="rbf"), param_grid, cv=5, scoring='accuracy')
grid.fit(X_train_sc, y_train)
print("Best params:", grid.best_params_)
print("CV accuracy:", grid.best_score_)
print("Test accuracy:", grid.score(X_test_sc, y_test))
Best params: {'C': np.float64(0.1), 'gamma': np.float64(10.0)}
CV accuracy: 0.7416576576576576
Test accuracy: 0.75
Exercise 9: Neural Networks#
In this exercise, you will use tensorflow to create a single layer neural network to classify handwritten numbers from 0 to 9 from the MNIST dataset.
Hint: Tensorflow is one of the most widely used machine learning learning libraries. It was initially developed by Google, but is open source and available for everyone. Tensorflow requires Python <=3.12. If you have an environment with Python 3.13, you either need to create a new one or simply use Google Colab for this exercise.
from tensorflow.keras.datasets import mnist
import matplotlib.pyplot as plt
# Load data and plot examples
(x_train, y_train), (x_test, y_test) = mnist.load_data()
fig, ax = plt.subplots(1,5)
for i in range(5):
ax[i].imshow(x_train[i], cmap='gray')
ax[i].set_axis_off()
plt.show()
We can then create the network with the following characteristics:
Input: A flattened version of the MNIST image (a vector of size 784)
Architecture: A single dense (fully connected) layer with 10 neurons (one for each class)
Activation function:
softmax(outputs probabilities summing to 1)Output: A probability distribution over digits 0–9; the highest is chosen
Learning rule:
categorical_crossentropyloss and stochastic gradient descent (SGD) optimiserEvaluation metric:
accuracy, measuring the percentage of correctly classified images
Tasks:
Explore the code and try to understand what it does (change things and see how they affect the result!)
Improve the model to achieve a better predicion accuracy (>97%). Potential change you can make:
Change the number of epochs or batch size (the number of training examples processed at once before the model weights are updated)
Change the learning rate or optimiser (use e.g. Adam, which uses an adaptive learning rate and is faster)
Change the model structure by e.g. adding a hidden layer with e.g. 64 or 128 neurons and a ReLu activation function
Compare your model with other students. Who managed to get the highest testing accuracy?
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Input, Dense
from tensorflow.keras.utils import to_categorical
from tensorflow.keras.optimizers import SGD, Adam
# 1) Load and preprocess data (flatten & scale)
(x_train, y_train), (x_test, y_test) = mnist.load_data()
x_train = x_train.reshape(-1, 28*28).astype('float32') / 255.0
x_test = x_test.reshape(-1, 28*28).astype('float32') / 255.0
y_train = to_categorical(y_train, 10)
y_test = to_categorical(y_test, 10)
# 2) Create the model: One dense (fully connected) with a softmax activation function
model = Sequential([
Input(shape=(784,)),
Dense(128, activation='relu'),
Dense(10, activation='softmax')
])
# 3) Compile & train the model
model.compile(loss='categorical_crossentropy',
optimizer=Adam(),
metrics=['accuracy'])
model.fit(x_train, y_train, epochs=5, batch_size=32, verbose=1)
# 4) Evaluate the model
loss, acc = model.evaluate(x_test, y_test, verbose=0)
print(f"Test accuracy: {acc:.4f}")
Epoch 1/5
1875/1875 ━━━━━━━━━━━━━━━━━━━━ 2s 1ms/step - accuracy: 0.8805 - loss: 0.4320
Epoch 2/5
1875/1875 ━━━━━━━━━━━━━━━━━━━━ 2s 1ms/step - accuracy: 0.9646 - loss: 0.1205
Epoch 3/5
1875/1875 ━━━━━━━━━━━━━━━━━━━━ 2s 995us/step - accuracy: 0.9770 - loss: 0.0782
Epoch 4/5
1875/1875 ━━━━━━━━━━━━━━━━━━━━ 2s 1ms/step - accuracy: 0.9828 - loss: 0.0564
Epoch 5/5
1875/1875 ━━━━━━━━━━━━━━━━━━━━ 2s 1ms/step - accuracy: 0.9869 - loss: 0.0420
Test accuracy: 0.9761
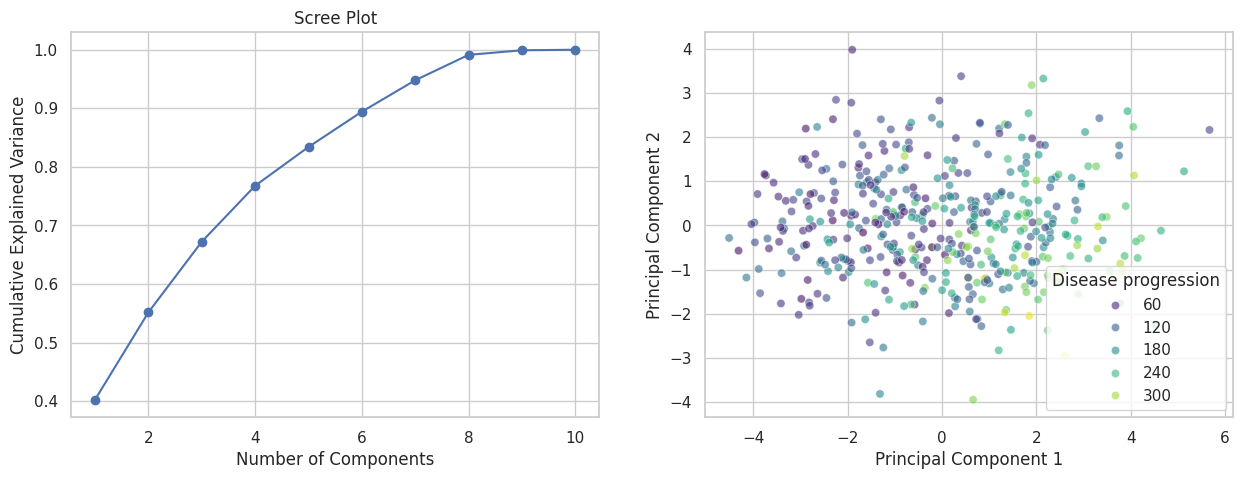
Exercise 10: Principal Component Analysis#
For today’s practical session, we will work with the Diabetes dataset built into scikit-learn. This dataset contains medical information from 442 diabetes patients:
Features (X): 10 baseline variables (age, sex, BMI, average blood pressure, and six blood serum measures).
Target (y): a quantitative measure of disease progression one year after baseline.
You can read more here: https://scikit-learn.org/stable/modules/generated/sklearn.datasets.load_diabetes.html
Tasks:
Inspect & clean (already implemented)
Display summary statistics (
df.describe()) for all 10 features.Check for missing values. (Hint: this dataset has none, but verify.)
Standardize
Use
StandardScaler()to transform each feature to mean 0, variance 1.
PCA & scree plot
Fit
PCA()to the standardized feature matrix.Plot the explained variance ratio for each principal component (a scree plot).
Decide how many components to retain (e.g.\ cumulative variance ≥ 80%).
Interpret loadings
Examine
pca.components_.For the first two retained PCs, list the top 3 features by absolute loading.
Infer what physiological patterns these components might represent.
Project the data for visualization
Compute the PCA projection:
X_pca = pca.transform(X_std).
Plot the results (already implemented)
Create a 2D scatter of PC1 vs. PC2, coloring points by whether the target is above or below the median progression value.
Do patients with more rapid progression cluster differently?
from sklearn.datasets import load_diabetes
# Load the data as a DataFrame
diabetes = load_diabetes(as_frame=True)
df = diabetes.frame
df.rename(columns={'target': 'Disease progression'}, inplace=True)
X = df.drop(columns='Disease progression')
y = df['Disease progression']
# 1. Inspect the data
df.head()
| age | sex | bmi | bp | s1 | s2 | s3 | s4 | s5 | s6 | Disease progression | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.038076 | 0.050680 | 0.061696 | 0.021872 | -0.044223 | -0.034821 | -0.043401 | -0.002592 | 0.019907 | -0.017646 | 151.0 |
| 1 | -0.001882 | -0.044642 | -0.051474 | -0.026328 | -0.008449 | -0.019163 | 0.074412 | -0.039493 | -0.068332 | -0.092204 | 75.0 |
| 2 | 0.085299 | 0.050680 | 0.044451 | -0.005670 | -0.045599 | -0.034194 | -0.032356 | -0.002592 | 0.002861 | -0.025930 | 141.0 |
| 3 | -0.089063 | -0.044642 | -0.011595 | -0.036656 | 0.012191 | 0.024991 | -0.036038 | 0.034309 | 0.022688 | -0.009362 | 206.0 |
| 4 | 0.005383 | -0.044642 | -0.036385 | 0.021872 | 0.003935 | 0.015596 | 0.008142 | -0.002592 | -0.031988 | -0.046641 | 135.0 |
import numpy as np
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
import seaborn as sns
sns.set_theme(style="whitegrid")
# 1. Standardize the data
scaler = StandardScaler()
X_std = scaler.fit_transform(X)
# 2. Perform the PCA
pca = PCA()
pca.fit(X_std)
# 3. Get the explained variance ratio
explained_variance = pca.explained_variance_ratio_
# 4. Project into PCA space
X_pca = pca.transform(X_std)
# 5. Plot the explained variance and 2D PCA projection
fig, ax = plt.subplots(1,2, figsize=(15, 5))
ax[0].plot(np.arange(1, len(explained_variance)+1), explained_variance.cumsum(), marker='o')
ax[0].set(xlabel='Number of Components', ylabel='Cumulative Explained Variance', title='Scree Plot')
sns.scatterplot(x=X_pca[:, 0], y=X_pca[:, 1], hue=y, palette='viridis', alpha=0.6, ax=ax[1])
ax[1].set(xlabel='Principal Component 1', ylabel='Principal Component 2');